Reconstruct Intermediate Sequences¶
Dowser automatically reconstructs intermediate sequences as part of the getTrees function. These are stored in the nodes list contained in each phylo object.
First, collapse internal nodes with identical sequences using the collapesNodes. This will significantly clean up the visualization. You could alternatively run getTrees with collapse=TRUE. Then, visualize the trees using plotTrees but with the node_nums parameter set. This will display the ID number of each internal node.
To obtain the IMGT-gapped sequence for each reconstructed node, specify the clone ID and node number in the getNodeSeq function.
library(dowser)
data(ExampleClones)
# Collapse nodes with identical sequences. This will
trees = collapseNodes(ExampleClones[1:2,])
# Plot trees with node ID numbers
plots = plotTrees(trees, tips="c_call", tipsize=2, node_nums=TRUE, labelsize=7)
plots[[1]]
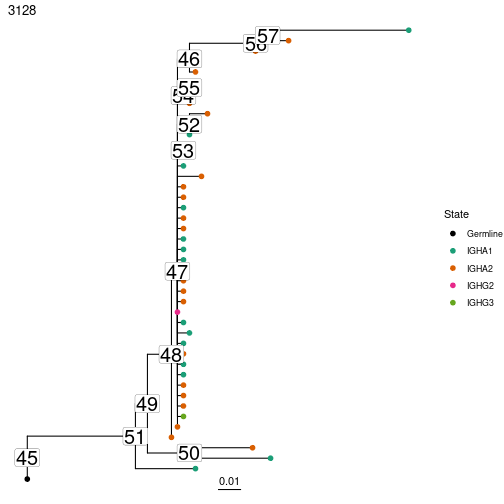
sequence = getNodeSeq(trees, node=50, clone=3128)
print(sequence)
## N
## "GAGGTGCAGCTGGTGGAGTCTGGGGGA...GGCTTGGTVCAGCCTGGGGGGTCCCTGAGACTCTCCTGTGCAGCCTCTGGATTCACCTTC............AGTGAYTATGCCATGAGCTGGTTCCGCCAGGCTCCAGGGAAGGGKCTGGAGTGGGTGGGTTTCATTAGAAGCAGACGTTTTGGTGGGACGCCGGACTACGCCGCGTCAGTGAGA...GACAGATTCACCATTTCAAGAGACGATTCCAAAAGCATCGCCTATCTGCAAATGAACAGCCTGAAAACCGAGGACACAGCCGTGTATTTTTGTAGTAGAGATCTCGCGGTTATATCCACAATAGCTGGTACTAACTGGTTCGACCCCAGGGGCCAGGGAGCCCTGGTCACCGTCTCCTCAGNN"